

 Skip to navigation
Skip to navigation
Site Primary Navigation:
- About SDSC
- Services
- Support
- Research & Development
- Education & Training
- News & Events
Search The Site:

Published April 28, 2023
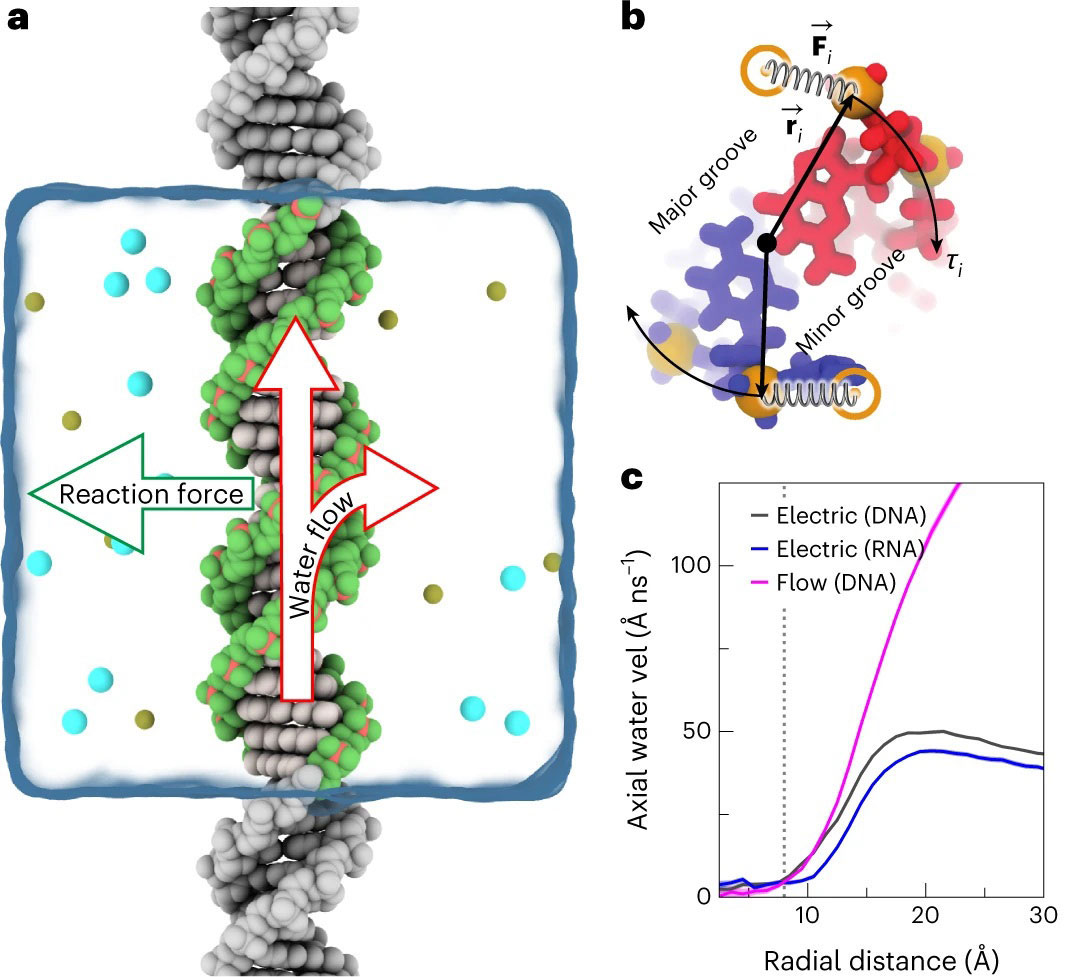
(a) Schematics of the simulation system in which the DNA was made effectively infinite by connecting its stands over the periodic boundary. The arrows illustrate the torque generation mechanism: redirection of the water flow at the DNA surface. (b) An illustration of a method used to measure or apply torque. A cross-section of the DNA construct is shown. Credit: University of Illinois at Urbana Champagne
By Kimberly Mann Bruch
For more than five years, researchers at the University of Illinois at Urbana Champagne have been studying how to create some of the tiniest motors ever invented – with inspiration from biological systems like DNA. The scientists recently used Expanse at the San Diego Supercomputer Center (SDSC) at UC San Diego to create simulations of the torque used by DNA molecules.
“Building things at this detailed nanoscale is very difficult in a lab, so we are using simulations of DNA molecules to see if they can act as the tiniest motors ever made,” said University of Illinois Professor Aleksei Aksimentiev, principal investigator for the study, which was recently published in Nature Nanotechnology.
Aksimentiev said that the first step in creating the Expanse simulations was to place the DNA molecules into water, apply electric field flow and then observe the rotation or measure the torque generated. Next, the team analyzed the simulations. They were most surprised by the magnitude of the torque, which was sufficient to produce rotation of larger molecular loads.
“People have thought about this for a while and gone back and forth—could it work or not,” Aksimentiev said. “We thought that maybe the water doesn't have enough traction to produce torque, but it did, and we were excited to see the DNA spin in the direction prescribed by its helicity despite significant fluctuations.”
Rotation of a DNA T-bar construct (with no external restrains) in a carbon nanopore. The top and side views are shown simultaneous in the two panels. Credit: University of Illinois at Urbana Champagne
A typical simulation on Expanse took a little over a week to complete—allowing the researchers to carefully create their simulations—measuring the torque and allowing them to see the rotation. Once they carefully analyzed their results, they submitted their work to be published in Nature Nanotechnology so that the broader research community has access to their findings.
“We continue to be interested in exploring how far we can push technology to build synthetic molecular motors that reproduce this simulation,” Aksimentiev said. “This work used only one DNA duplex and we will continue to look at other designs to further the research.”
Eventually, Aksimentiev said that he and the Illinois team plan to use lessons learned from this study to build systems at a nanoscale level that will be used as components in self-propelled systems or for nanoscale energy conversion.
This is no small feat, but with the help of Expanse, Aksimentiev feels that they can achieve their future research goals.
“Supercomputers like SDSC's Expanse are essential to the development of nanotechnology as they provide a window into the nanoscale world that is otherwise not accessible to experiments,” Aksimentiev said.
Computational work used the National Science Foundation Extreme Science and Engineering Discovery Environment (allocation MCA05S028).
Share